Dissemination / News
2020-02-27 WP3 progress report
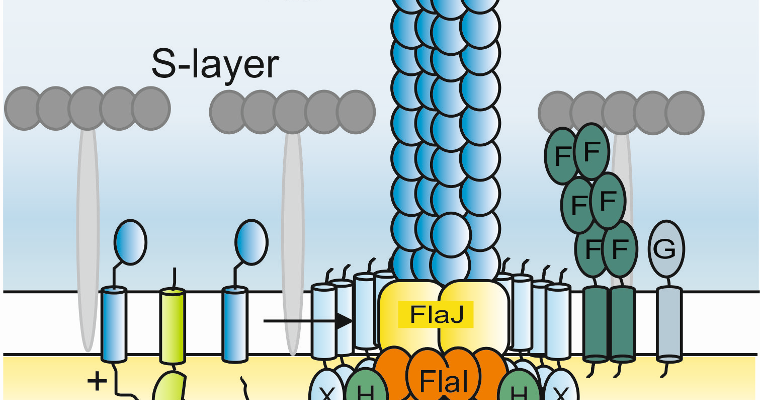
Imperial College and Freiburg have been developing a productive investigation into one of the most interesting aspects
of the mechanism of the archaellar motor: the stator complex. The archaellum is functionally analogous to the unrelated
bacterial flagellum and eukaryotic cilium. To rotate, the archaellar motor must anchor to the cell superstructure, and
evolution of this anchor will have been central to its development as a rotary motor. We have recently published an important
study in Nature Microbiology detailing the structure and function of the archaellar stator complex, and are now developing cryoEM
approaches to determine the near-atomic resolution structure of this component. We have also just submitted a substantial
authoritative review on the mechanisms and evolution of archaella, flagella, and cilia to FEMS Microbiology Reviews for
publication in the coming months.
Aarhus University has been researching the possibility of employing a DNA-based nanopore for self-insertion into a cell membrane.
The prediction of DNA-origami folds has seen vast improvements over the last few years and are now routinely being used for generating
semi-complex 3D structures. A DNA-based nanopore could be used to disrupt the natural salt ion balance of targeted cells and ultimately
kill them, while better avoiding an immune response than a naturally occurring protein ion-channel. The DNA nanopore is being studied by
electrophoresis measurements using Xenopus laevis oocytes as a model cell system, where even minor changes in the salt balance and subsequent
membrane potential can be readily detected. To improve the insertion process, conjugate chemistry has been used to produce DNA-peptides,
fusing a cell-penetrating peptide to the DNA pore. If successful, the DNA-pore could be integrated in the MORO system, as the cell-lysing component.
2020-02-21 WP4 progress report - MORO development
Task 5:3 Optimisation and evaluation of MORO functionality (AIT, ICL, AU)
To create the D-SEPEC based on the catalytic centre of the FlaI protein complex of the archaellum, a structurally
well-defined multi-complement DNA origami-based rotary device was constructed. The DNA origami rotor was fully
characterized by electron microscopy under negative stain and cryo preparations. We further analyzed the Brownian
and flow-driven rotational dynamics of the rotor in real time by single-molecule microscopy in a microfluidic
chamber set-up. The videos captured 800-2000 frames with an exposure time of 10 ms. Thunderstorm, a plugin for
image J for data analysis of super-resolution data, was used to localize the position of the propeller-attached
fluorophores in each frame of the video. From the rotor geometry and EM structures, we predict that the rotor can
rotate freely between three preferred rotational states. In this regard, the localization patterns of the nanomachines
ranged from a radially symmetric, to elongated, to triangular; we speculate that the latter is indicative of a rotor rotating
freely between the three rotational states. In addition, we observed a higher degree of mobility in the fully assembled
nanomachine as compared to the surface-immobilized rotor and propellers, and this mobility increased slightly under moderate flow,
showing the possibility to manipulate the dynamics of an artificial nanodevice with fluidic flow as a natural force.
2020-02-21 New publication in Nature Microbiology
The structure of the periplasmic FlaG-FlaF complex and its essential role for archaellar swimming motility.
Tsai C.-L., Tripp P, Sivabalasarma S, Zhang C, Rodriguez-Franco M, Wipfler RL, Chaudhury P, Banerjee A, Beeby M, Whitaker RJ, Tainer JA, Albers SV.
2020. Nature Microbiol, 5(1):216–225.
2020-02-12 WP2 progress report
As the MARA project reaches its final phases the advancements in all the work packages
amass. The WP2 comprises the (1) selection of chemically modified aptamers by Pure
Biologics which are meant to recognize important bacterial pathogens and particularly
certain proteins responsible for the antibiotic resistance of microorganisms, as well as (2)
development of the ADUENAs – AUtonomous DEtection Nucleic Acids, jointly by Austrian
Institute of Technology (AIT) and Pure Biologics. Currently a package of several already
promising aptamers, recognizing a set of selected bacterial targets, is being thoroughly
characterized and optimized to enable refinement of candidates for diagnostic and targeting
molecules. Incorporation of those aptamer binders has also already been initially tested as
part of the AUDENAs.
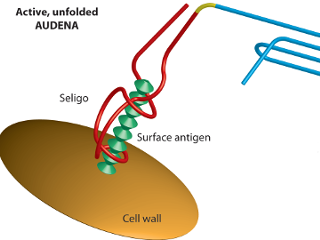
In the same time, AIT team is working on the design and experimental analysis of AUDENAs. The
DNA G-quadruplex (GQ) is used as the reporter system in the AUDENA. The hemin/GQ complex
mimics the catalytic activity of the HRP enzyme and catalyzes the peroxidase reaction of
ABTS/H2O2 generating a colorimetric signal. In addition, GQ can act also as a light-up aptamer
and binds to Thioflavin T forming a fluorescent complex. Four different aptamers targeting protein A,
staphylococcus enterotoxin B, ATP and quinine were selected from the literature. Despite designing ca.
200 AUDENA probes using distinctive design principles followed by extensive experimental analysis using
two independent techniques (colorimetric and fluorescence assay), only few of those AUDENA probes displayed
functionality. Using ELONA (Enzyme Linked Oligonucleotide Assay), we found that the affinity of the integrated
aptamer is dramatically reduced or even lost upon insertion into the AUDENA. Even adding a G-quadruplex
sequence (e.g. PS2M) to an aptamer can adversely affect the affinity. The lack of target-induced
structural switching functionality in the chosen aptamer also contributes to the low functionality.
Taking advantage of split aptamers and split G-quadruplex, we have been able to build functional AUDENAs.
Both ATP and quinine-binding aptamers are engineered into a split-aptamer comprising two nucleic acid strands
that co-assemble as they bind to their target. By adding the split G-quadruplex to each split aptamer, we
were able to design a functional AUDENA.
2019-12-02 8th General Assembly Meeting
The 8th MARA project General Assembly meeting took place in Vienna on December 2nd and 3rd, 2019.
2019-11-19 TV report on Pure Biologics and the MARA project
We are happy to share a TV material from TVP Wroclaw, a Polish regional TV broadcast, discussing Horizon 2020 funding and the participation of Pure Biologics in the MARA project! Pure Biologics’ experts, Dr. Agnieszka Sok-Grochowska and Monika Czarnecka answer a couple of questions regarding the projects scope and aims.
2019-05-14 7th General Assembly Meeting
The MARA project 7th General Assembly Meeting has been held at Pure Biologics’ site in Wroclaw, Poland, on 14th-15th May. Pure Biologics was pleased to welcome all the consortium partners and discuss recent advancements in the project as well as future plans for collaboration and research!
2019-02-14 WP4 progress report – Creation of D-SEPECs
DNA Scaffold Embedded Protein Emulation Complexes – or for short D-SEPECs – are self-assembling nanostructures which
have artificial catalytic centres emulating existing proteins embedded within the scaffold.
A proof-of-concept D-SEPEC is planned to be developed during Work Package 4 of the MARA project.
Scientists working within the MARA project at the AIT Austrian Institute of Technology have developed the software ADENITA
for the design of complex DNA nanostructures. Some highlights of this tool are that proteins can be embedded in the designs
and that the designs are available on an atomic scale. A public version will be released in February, and the first structures
designed with ADENITA are currently being visualised in vitro. To that end, Imperial College London is working to image DNA origami
constructs using electron microscopy and electron cryo-microscopy to verify correct assembly and to monitor the dynamics of
these structures. The team has already successfully imaged a number of structures in 2-D, and are now working to determine
3-D structures.
In the same time at AIT, first artificial DNA-based enzymes – the D-SEPECs – are undergoing rigorous trails to
confirm the already-observed effect of the hydrolysis of ATP, which is one of the most important chemical reaction catalysed by enzymes.

2019-01-21 New JOVE publication
2018-12-18 6th General Assembly Meeting
The 6th MARA project General Assembly meeting took place in London on December 18th and 19th, 2018.
2018-10-08 WP3 progress report
The contribution of Work Package 3 is to determine how a set of naturally-occurring molecular machines work;
this understanding will ultimately form the basis for designing the MORO for the MARA project.
As such, Work Package 3 is a set of different but inter-related research projects by the consortium of Imperial College London,
Albert-Ludwigs-Universität Freiburg, and Aarhus University.
Together, Imperial College and Albert-Ludwigs-Universität Freiburg have been working to understand how the molecular propeller
used by exotic “archaea“ microbes operates. This ongoing collaboration has led to development of a variety of methodologies for
optimal growth of two archaea, Sulfolobus acidocaldarius and Haloferax volcanii. Work is now underway to use electron cryo-tomography
to directly visualize the functional molecular propeller in the cell. If successful, these efforts will guide the design of the
rotary component of the MORO. Imperial College and Albert-Ludwigs-Universität Freiburg are also completing a comprehensive
literary review of molecular propellers across all life on earth, to be published in the coming months.
At the same time, Aarhus University have been researching the mechanism of an “ion channel” that could be used in the MORO
to attack cancer cells or invading pathogens. The insertion of such an ion channel into the cell membrane will disrupt the
natural salt ion balance and ultimately destroy the targeted cell. The ion channel has been expressed and purified from
mammalian cells and is being investigated by state-of-the-art electron microscopy, to study its ion selectivity mechanism.
The recent emergence of DNA-based nanopores, which can be inserted into biological membranes and maintain ion selectivity
are also being investigated. These could potentially be integrated in the MORO and the entire complex is to be analysed by
electron cryo-tomography resulting in 4D-models of a mobile cell lysing nanobot.
2018-10-02 ArchaeaBot
ArchaeaBot is an underwater robotic installation by Anna Dumitriu and Alex May that explores what ‘life’ might mean in a
post-singularity and post-climate change future. The project is being supported through a EMAP/EMARE artists residency at
LABoral Centro de Arte y Creación Industrial in Spain via funding from Creative Europe and with generous support from Arts
Council England.
The artists have based their project on research funded by MARA; specifically the work on archaea being carried out at Imperial College
London and Albert-Ludwigs-Universität Freiburg, combined with the latest innovations in artificial intelligence and machine
learning, in order to create a hypothetical hybrid species for the end of the world.
The work is the result of collaboration with Amanda Wilson (Research Associate. Imperial College London) within the framework of the EU MARA
project, and Daniel Polani (Professor of Artificial Intelligence in the School of Computer Science at the University of
Hertfordshire).
ArchaeaBot is a motorised swimming robot, designed to emulate a Sulfolobus acidocaldarius cell, the model organism being used to investigate
the structure of the archaellar motor. The robot is roughly spherical, and embedded in the surface are several 3D-printed
computerised motors which drive the rotation of flexible filaments, in order to simulate archaellar motors and their archaella.
The artists were inspired by the knowledge that Sulfolobus acidocaldarius thrives in a hot acid environment, and came up with the notion that
theoretically human consciousness could be uploaded to these extremophiles in order to survive the hot acid conditions that
may prevail on Earth in the future. Of course, this goes well beyond the confines of the original MARA research, but the
artist’s role is to dream and work outside the normal boundaries, and the story draws people in, so they want to find out
more about the science.
ArchaeaBot received critical acclaim when it was exhibited at the prestigious Ars Electronica Festival 2018, an annual gathering of of
artists, scientists and technologists, intended as a setting for experimentation, evaluation and reinvention. The artwork
was also exhibited at the LABoral Centre for Art and Industrial Creativity, following a discussion between Anna Dumitriu,
Alex May, Amanda Wilson and Institute director, Karin Ohlenschlaeger to a live audience, and will soon be presented at Art
& the Life Sciences: Ethics & Perspectives in Athens, Greece.
ArchaeaBot will be further developed over the next year, with an aim to add a translucent S-layer, refined artificial motors and interactive
features. The artists also hope to build a second robot based on the other model organism that is being used for structural
electron cryo-microscopy investigations; Haloferax volcanii.
From the MARA perspective, the project is achieving excellent outreach, helping to inform the public about the MARA objectives and the biology
of Archaea.
Image credit: Vanessa Graf - Ars Electronica
2018-09-19 Computational DNA engineering workshop
From Sep, 17th to Sep 19th members of the MARA project participated in the Computational DNA engineering workshop in Plitvice, Croatia. Within this international and highly successful meeting researchers from different computational areas presented their work and discussed possible collaboration opportunities. The diverse research backgrounds and domain knowledge of the workshop participants led to fruitful discussions about relevant research questions, problems and ideas.
2018-06-14 WP2 progress report
The Work Package 2 comprises the development of modified aptamers recognizing several important bacterial pathogens, the design and testing of model
Autonomous Detection Nucleic Acids (AUDENAs) to achieve a pathogen detection tool with exchangeable specificity-determining unit.
The tasks are handled by Austrian Institute of Technology (AIT) and Pure Biologics Inc. (PB, who replaced Apta Biosciences
as the consortium partner in late 2017).
Recently, Pure Biologics has finished the first package of selections directed towards chosen bacterial targets and started another set of selections.
The results are currently being analysed using Next Generation Sequencing techniques, which in conjunction with powerful bioinformatical analysis
allow to readily identify multiple potential binders of the bacterial targets. Next, identified aptamers will be tested and optimized in order to
incorporate them into working AUDENAs.
Parallelly, scientists at AIT have designed various AUDENA concepts and tested the new sensor class with previously published conventional
DNA aptamers. With these experiments they could proof that the AUDENA technology can be used for the detection of bacterial antigens.
A publication of the results in a scientific journal is planned in 2018 to demonstrate the usability of these cost-effective, stable and
point-of-care compatible detection molecules.

2018-05-14 5th General Assembly Meeting
The 5th MARA project General Assembly meeting took place in Freibug on May 14th and 15th, 2018.
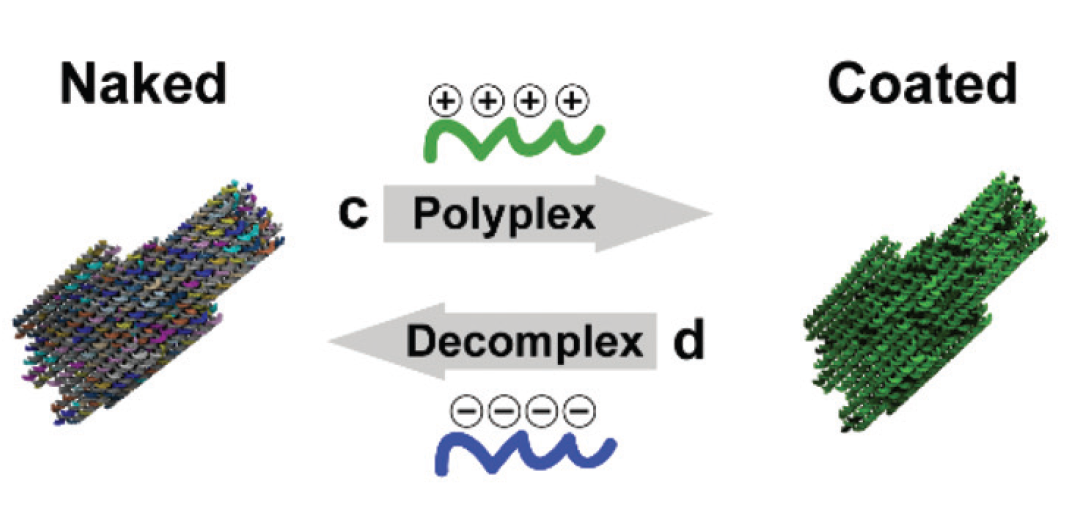
2018-03-20 New publication in Nanoscale
(Poly)cation-induced protection of conventional and wireframe DNA origami nanostructures.
Yasaman Ahmadi, Elisa De Llanoa, Ivan Barišić. Nanoscale. 2018 Apr 26;10(16):7494-7504.
2017-12-30 Pure Biologics joins consortium
The Polish company Pure biologics joined the consortium in December 2017, as Apta Biosciences unfortunately left the project.
2017-12-19 4th General Assembly Meeting
The MARA project GA meeting took place in Vienna, on December 19th, 2017.
The next meeting will be in Freiburg on May 14th and 15th, 2018.
2017-08-29 New publication in IEEE Trans Vis Comput Graph
Multiscale Visualization and Scale-Adaptive Modification of DNA Nanostructures.
Miao H, De Llano E, Sorger J, Ahmadi Y, Kekic T, Isenberg T, Groller ME, Barisic I, Viola I. IEEE Trans Vis Comput Graph. 2018 Jan;24.
2017-03-23 3rd General Assembly Meeting
The MARA project GA meeting took place in Aarhus, on May 23rd and 24th, 2017.
2016-06-02 2nd General Assembly Meeting
The MARA project General Assembly meeting took place in London, on June 2nd and 3rd, 2016.
2016-11-24 1st General Assembly Meeting
The 2nd MARA project General Assembly meeting took place in Slunj, on Nov 24th and 25th, 2016.
2016-03-30 News article
News article from Die Presse about the MARA project.
2016-01-29 News article
News article from Die Presse about the MARA project.
2016-01-29 News article
News article from Chemie report about the MARA project.
2016-01-28 Very Important Business
MARA news on "Very Important Business".
2016-01-28 Press release
MARA is in the news. Please click on the link to read the full press release.
2015-11-10 Kickoff Meeting
The MARA project kick-off meeting took place in Vienna, Austria, on December 10th and 11th 2015.
2015-11-04 Apta Biosciences news
Apta Biosciences reports about the newly granted MARA project.